TPR Reviews IBM’s PatCID – TPR has been working with the IBM Deep Search research team to evaluate their patent chemical structure database and search tool, PatCID. We’ve written about this previously, and what follows is our March 2025 update to this series.
Previous articles on this topic are available on Technology & Patent Research (TPR) Insights; Part 1 and Part 2.
Introduction to PatCID
PatCID (Patent-extracted Chemical-structure Images database for Discovery) is a chemical structure search tool developed by IBM’s Deep Search research team in Zurich, Switzerland. The name refers both to the sequence of algorithms used to process patent documents and identify unique 2D chemical structures, as well as the resulting dataset.
While the most commonly used chemical structure database tools excel at indexing example structures and structures found in claims, many structures embedded within patent documents—often spanning hundreds of pages—may not be directly searchable in these popular resources. PatCID addresses this gap by automatically indexing these often-overlooked structures, increasing coverage and improving the likelihood of discovering relevant prior art.
Beyond patent literature, PatCID’s processing pipeline can also be applied to personal PDF collections, streamlining workflows and accelerating discovery efforts for researchers and intellectual property professionals.
How PatCID Works
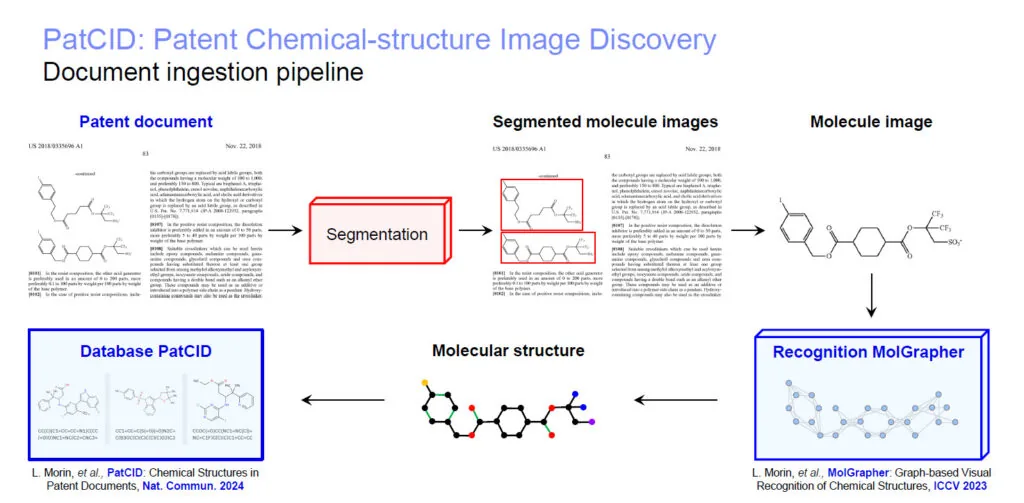
PatCID uses a sophisticated processing pipeline to identify and categorize chemical structures:
- Segmentation Module: This module identifies chemical structure images within documents.
- Molecule Classifier: The identified images are categorized based on their chemical content.
- Molecule Grapher: The structures are converted into SMILES (Simplified Molecular Input Line Entry System) format, which are then stored in the PatCID database.
The IBM Deep Search team has highlighted improvements to their segmentation module, significantly enhancing the identification and separation of chemical structure images, such as those found in charts or lists. They have also improved their molecule classifier, which better differentiates 2D molecular representations that can be successfully graphed from Markush structures.
[Image courtesy of IBM Deep Search research team]
PatCID’s initial dataset (V1.0) is available on GitHub and includes chemical data extracted from organic chemistry patent publications using the keyword “alkyl.” The dataset covers patents from as early as 1978 through 2022 (coverage varied by patent authority, see Morin et al Nature Communications article, Table 1) and includes:
- USPTO: 0.7M documents
- JPO: 1.0M documents
- CNIPA: 0.6M documents
- KIPO: 0.4M documents
- EPO: 0.4M documents
This dataset includes 3.1 million documents and 13 million unique structures. According to IBM’s Deep Search’s comparisons with SciFinder and Reaxys, there is significant overlap, while PatCID also captures some structures missed by both1. However, many structures found in SciFinder and Reaxys are not present in PatCID.
Expanded Coverage in V2
Since our testing began, the IBM Deep Search team has significantly expanded document coverage. The date range was extended back to 1980. The team is also updating their coverage of recently available IP5 and WIPO publications. Combined with further improvements to the segmentation module, this enabled the processing of 4.3 million documents in V2, resulting in a library of 24 million unique structures.
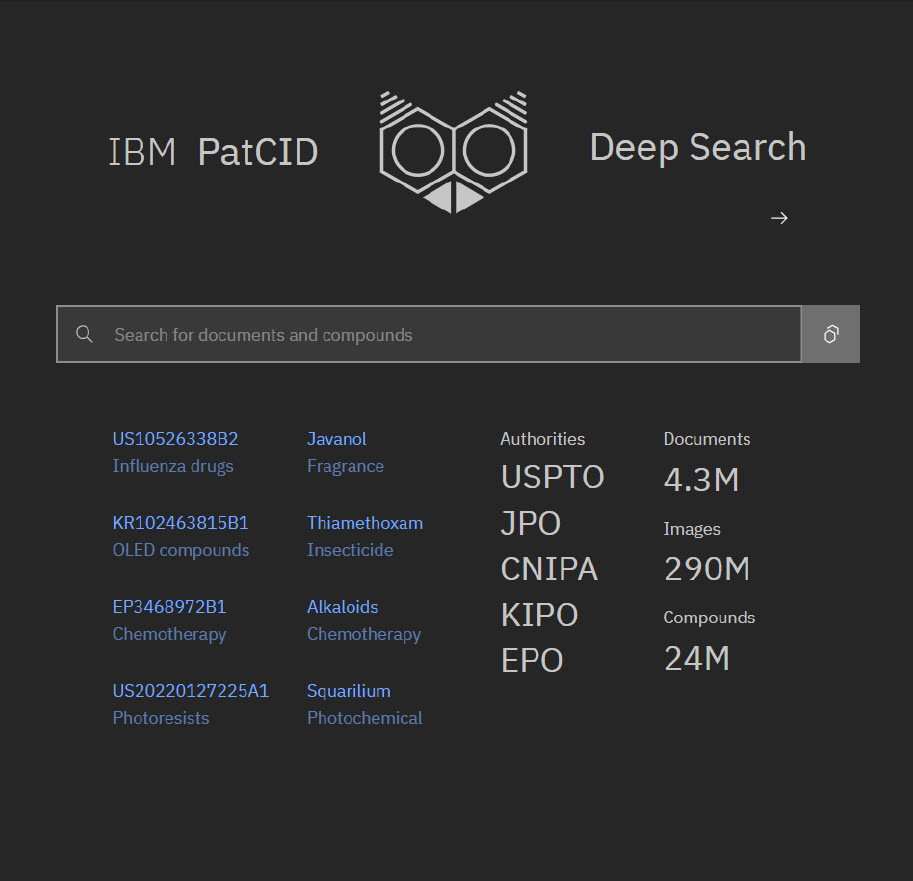
These updates have created a broader and more detailed chemical structure database, further establishing PatCID as a valuable resource for researchers and patent professionals.
TPR Testing Results and Observations
In our (TPR’s) initial testing, PatCID demonstrated particular strengths and some limitations:
- Exact Compound Searches: These are most successfully executed in PatCID, offering intuitively easy-to-navigate results.
- Substructure Search: While functional, the tool currently lacks mechanisms to adjust variability in structure queries (e.g., wildcard codes for “G” groups, cyclic or hydrophilic groups, variable attachment points).
- Similarity Search: Provides unique results in addition to CAS tools, offering additional perspectives in structure analysis.
- Missed Structures: PatCID has located structures missed by other tools but has also overlooked some indexed by alternatives like SciFinder and PubChem.
Below are some typical navigation views from within PatCID (provided with the permission of IBM’s Deep Search research team).
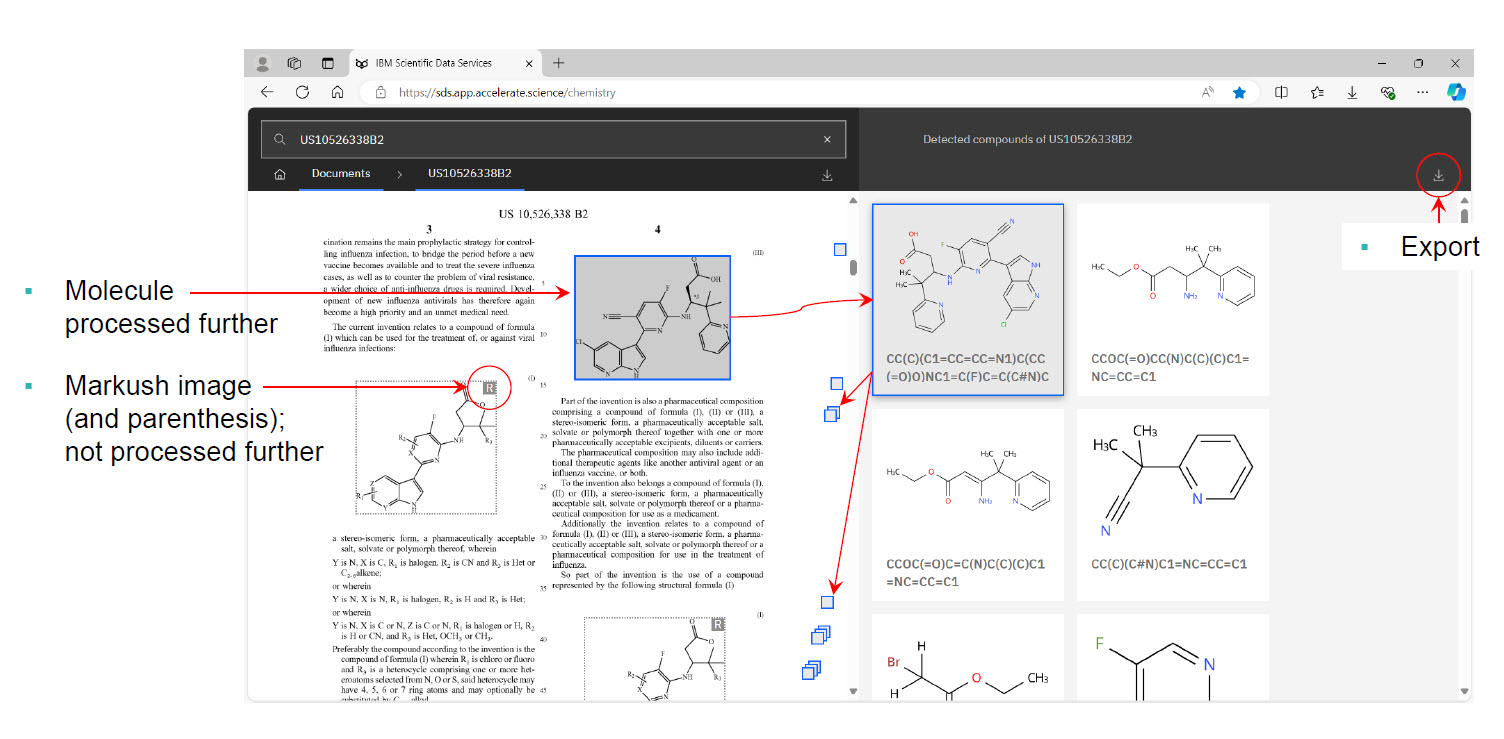
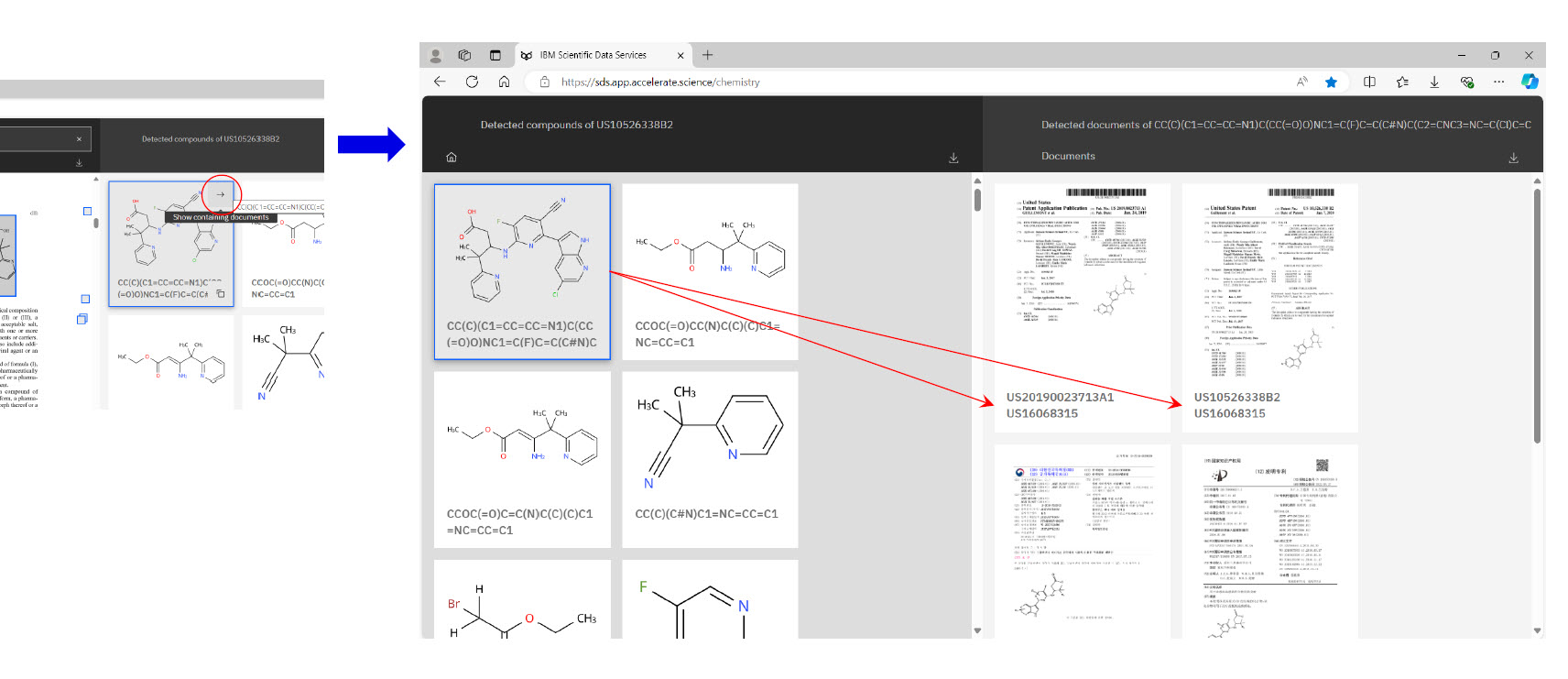
TPR Insights from Specific Test Cases
- Isotopic Structures: PatCID did not directly identify an 18F atom on a molecule, instead matching the non-isotopic fluorine atom. However, the tool highlighted both the isotopic and non-isotopic structures in the results. Discussions with IBM revealed that it can be trained to detect isotopic molecules, but this has not been prioritized at this time. Further from the Deep Search team: “Arguably the most relevant isotope would be the hydrogen deuterium. For PatCID v2.0 the Molecule Grapher was retrained to now also cover the hydrogen-isotope deuterium (which is displayed in the molecular-structure images as a D or 2H). PatCID V2.0 now also cover(s) deuterated molecules.”
- Keyword Dependency: A missed reference was traced to a Chinese patent publication that lacked the “alkyl” keyword, excluding it from processing. Due to the improved efficiency of the segmentation module, future updates are aimed to eliminate keyword dependency, enabling processing of all available publications. We have come to learn that this “alkyl” keyword limitation only applies to non-US patent documents, as all USPTO documents after 2001 have been processed for both PatCID V1.0 and V2.0.
Recent and Ongoing Updates to PATCID
Several significant improvements have been a recent focus:
- Vector Database Implementation: A new vector database would address issues with substructure searches, enabling more efficient and accurate results.
- Enhanced Query Control: Further query capabilities, using so-called SMARTS (SMILES arbitrary target specification), have been implemented, that provides users greater control over bond specifications and other structural nuances.
- Editor Updates: Some improvements to the structure editor are already applied, and more are planned to streamline the user experience.
Long-Term Updates (2025 and Beyond)
IBM has ambitious plans for PatCID’s future:
- Markush Structure Processing: Automated handling of some Markush structures using multimodal AI. This approach combines image and text-assessing algorithms, showing promising early results.
- Dataset Accessibility: While V1.0 data will remain free on GitHub, newer datasets with advanced algorithms will be available exclusively on the Deep Search platform, which is not free of charge.
Conclusion – TPR Reviews IBM’s PatCID
PatCID represents a significant advancement in chemical structure patent searching, providing expanded coverage, improved segmentation for automated indexing, and unique search results. While it has some limitations, ongoing updates and enhancements suggest it will become an increasingly valuable tool for researchers and patent professionals. For a comprehensive chemical structure search, PatCID should be considered for use alongside, rather than as a replacement for, other industry-standard chemical structure databases.
Stay tuned as TPR continues to explore and share our experiences with this evolving platform.
To complement this edition, where TPR Reviews IBM’s PatCID (March 2025), you can find additional insights in our earlier reviews in Part 1 and Part 2. Have questions regarding chemical patent searching and want to discuss with our team, please contact us.
References
- Morin, L., Weber, V., Meijer, G. I., Yu, F., & Staar, P. W. J. (2024). PatCID: an open-access dataset of chemical structures in patent documents. Nature Communications. Published on August 2, 2024. https://www.nature.com/articles/s41467-024-50779-y
© Technology & Patent Research International, Inc.


