In the previous blog (Part 1), we provided an initial overview of PatCID (IBM’s new open-access patent chemical structure database). In this blog (Part 2), we will expand on the paper’s content, share our notes from discussions with the IBM research team, and offer our impressions and takeaways about the database.
How PatCID Works
PatCID is built around three core components that work together to automate the classification and retrieval of chemical structures:
- DECIMER-Segmentation: This module identifies images of chemical structures in patent documents, “segmenting” them from the rest of the document.
- MolClassifier: Once identified, this component classifies structures into relevant categories, such as molecular or Markush structures (which can’t be properly graphed yet).
- MolGrapher: This component converts the images into molecular graphs, which can be stored as machine-readable formats like SMILES (Simplified Molecular Input Line Entry System).
The system is highly effective in recognizing molecules from recent patents and surpasses traditional rule-based models like OSRA and competes with state-of-the-art deep-learning models like DECIMER in both precision and recall. The database draws primarily from patent documents published by the USPTO, EPO, JPO, KIPO, and CNIPA between 2010 and 2022, although some patents date as far back as 1978. PatCID has indexed over 81 million chemical images and 14 million unique structures, which exceeds the coverage of competitors such as Google Patents and SureChEMBL.
PatCID’s focus is on organic chemistry patents, which are selected through keyword filtering with the term “*alkyl*”. The selection of patent offices was also strategic, as they account for 90% of the 1.16 million patent families in organic chemistry published worldwide as of January 2024, per investigation using TotalPatent One.
Users of PatCID can search the database by molecular similarity, substructure, or as-drawn queries. Additionally, they can navigate by molecule within the indexed documents, pinpointing the exact location of molecules within the patent text.
Precision and Recall in Context
To fully appreciate PatCID’s value, it’s essential to understand two critical performance metrics in information retrieval: precision and recall.

- Precision is the proportion of relevant items retrieved by a search query out of the total number of items returned by the same query. This is a measure of how many results are relevant.
- Recall is the proportion of relevant items retrieved by a search query out of the total relevant items in the dataset. In fields like drug discovery and materials science, high recall is vital—missing relevant chemical structures can hinder innovation or lead to overlooking important intellectual property.
Achieving high recall through automation is challenging. One major difficulty is the variability in patent formats; chemical structures can appear as text, images, or complex Markush structures. Additionally, geographical and chronological diversity in patent data adds complexity—older patents or those from certain regions, like Asia, may be poorly formatted or of lower quality, making retrieval harder.
Research and Testing of PatCID
IBM researchers tested PatCID against commonly used structure-searching tools, including fully automated indexes like SureChEMBL, and Google Patents, as well as databases which use a combination of automatic and manual indexing like Reaxys and SciFinder. To evaluate PatCID’s performance, two benchmark datasets were created:
- D2C-RND (Document to Chemical Structures – Random): This dataset was used to test how well PatCID handles newer patents from jurisdictions like the U.S., where patent formatting is generally consistent.
- D2C-UNI (Document to Chemical Structures – Uniform): This dataset evaluated PatCID’s performance on older patents and those from non-U.S. regions, which sometimes feature lower-quality or non-standard chemical structure images.
The Data
Several of the following tables and graphics are taken from the Morin et al paper, with some modification. Table 2 was generated by TPR.
Table 1 Patent databases statistics
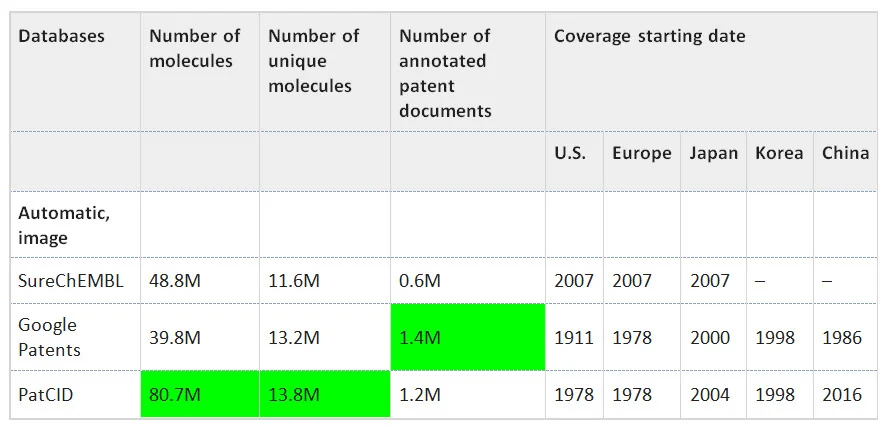
Number of patent documents, number of molecules, and number of unique molecules covered by patent databases. Which offices are covered and since when.
This table has been modified from the original version in two ways. Originally it contained rows for SciFinder and Reaxys, and we removed those lines as data is unavailable for the key fields of interest. Second, highlighting was applied to the cells containing the largest values for each feature column. In terms of the number of total and unique molecules indexed, PatCID contains a larger database total than the two other automatic indexing resources evaluated. While the total number of annotated patent documents is greatest for Google Patents, PatCID’s count is within 15% of the larger total.
Table 2 Figures not limited to the study conducted by the PatCID group
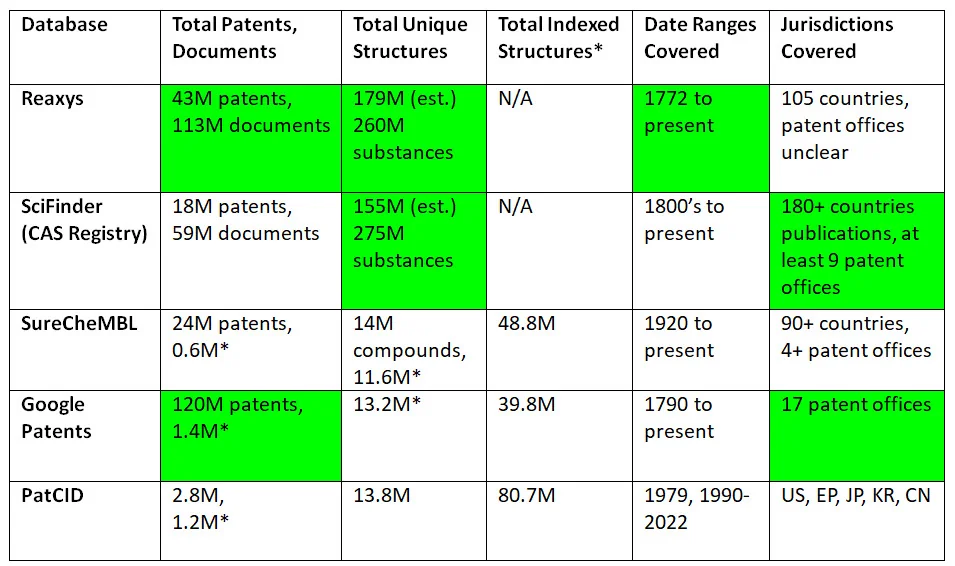
*Within the study scope – organic chemistry patents from corresponding date ranges and jurisdictions.
This table attempts to look more broadly at each database, instead of focusing on just patents within the jurisdictions and time frame of interest. These are not all official numbers – when the database providers made those readily available, they were incorporated into the table, but some of these numbers are best approximations based on publicly available information.
We compiled this table to illustrate and to emphasize that right now, while the PatCID patent chemical structure database is impressive within the parameters of its development and study, if exhaustive searching is necessary, other resources should also be included in the search strategy.
Table 3 Search comparison from the Morin et al paper (IBM) for automatically-created databases
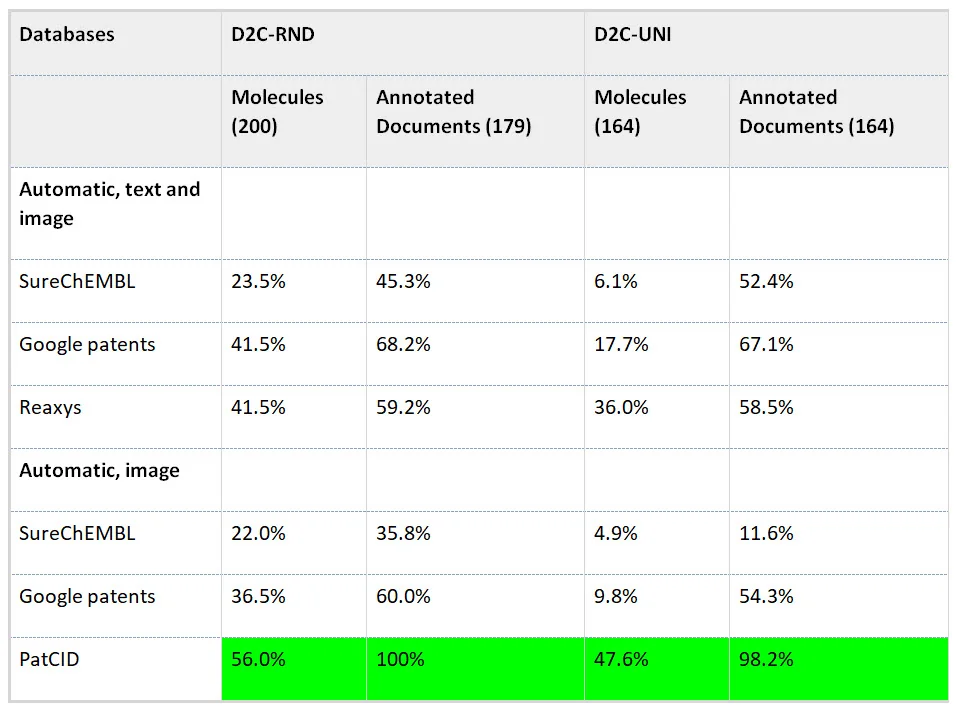
Comparison of the molecule and document retrieval performances of state-of-the-art automatically-created patent-databases. The recall of molecules and annotated documents is reported for benchmarks based on random (D2C-RND) and uniform (D2C-UNI) distributions of chemical images. The numbers in between parentheses are the numbers of samples in each set.
In this table, we made one modification: to highlight the largest value in each field.
When looking only at the automated molecule and document recall performance, for both benchmark datasets, PatCID outperforms all assessed databases. This is quite an encouraging result, especially the high marks for document recall in both sets.
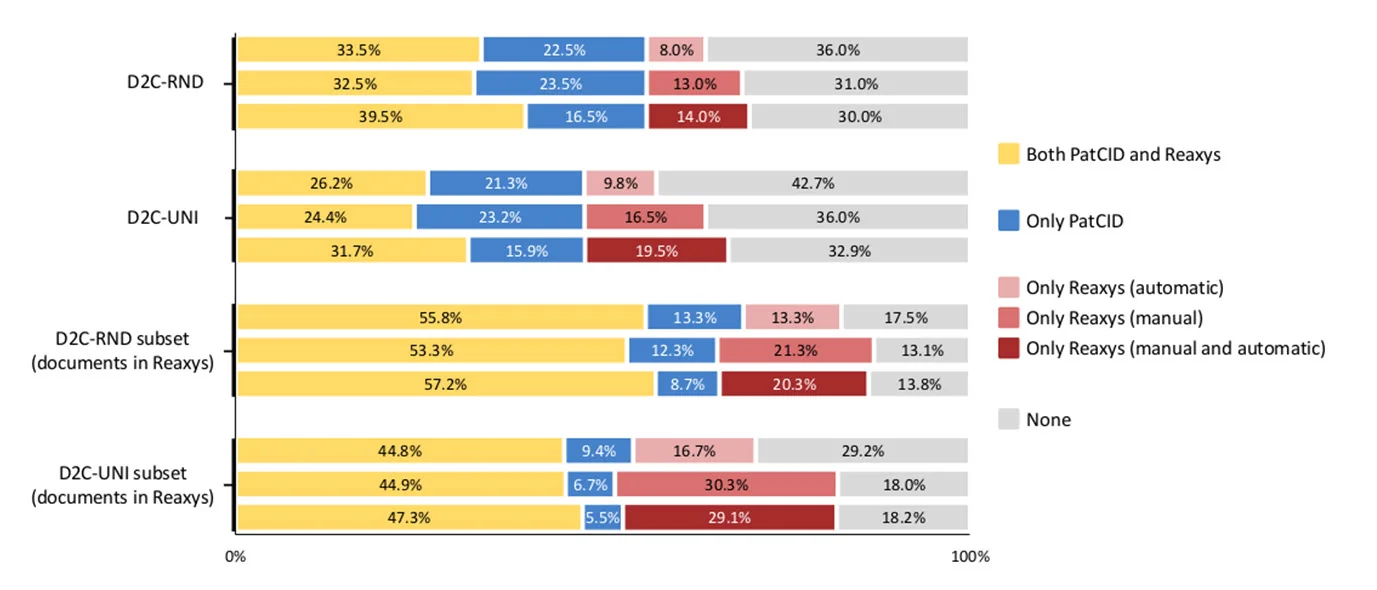
Figure 1 Search comparison between PatCID and Reaxys
Proportions of molecules covered in the PatCID and Reaxys databases for the random (D2C-RND) and uniform (D2C-UNI) benchmarks, and their subsets restricted to documents annotated in Reaxys. For the top bar, 33.5% of molecules in D2C-RND are covered with both Reaxys (automatic annotations) and PatCID, 22.5% in PatCID only, 8.0% in Reaxys (automatic annotations) only, and 36.0% in none of the databases.
This figure is an update to the original that appeared in Nature, with corrected legend. In the first print, the “Only Reaxys (automatic)” and “Only Reaxys (manual)” labels were reversed in error.
For each benchmark set, we see considerable overlap in the documents recalled by both PatCID and Reaxys. Broadly speaking, PatCID tended to recall a greater number of unique patents than Reaxys did, even when combined automated and manual indexing was accounted for – however there was some number of documents in the benchmark group which didn’t appear within Reaxys. When the comparison is adjusted to assess PatCID recall in only those documents which Reaxys also processed, the recall by Reaxys exceeds that of PatCID.
First Impressions and Initial Takeaways of PatCID
Although the full database of chemical structure indexes can be downloaded for free from the GitHub repository, TPR has been granted temporary access to the PatCID module on IBM’s Deep Search platform. From TPR’s initial testing of the system, PatCID database offers a user-friendly experience, with fast linking to other documents containing similarly tagged structures. Navigating between structures and patent documents is simple and intuitive. Exporting search results is easy, though currently, export files are limited to basic listings of patents. Future updates may include more detailed information, such as whether the chemical structure appears in the claims section of the patents.
However, there are some limitations to PatCID’s current coverage. It is restricted to patent documents, specific jurisdictions, a certain date range, and by the ‘*alkyl*’ keyword selection. Intellectual property practitioners need to be mindful of these gaps, especially in freedom-to-operate (FTO) and invalidity studies, where overlooking a critical patent could carry significant risk and cost.
That said, PatCID’s ease of use and relative reliability make it an excellent supplement to existing workflows. It seems to bridge the gap between free automated tools and more expensive manually-curated resources.
Looking Ahead
According to Dr. Meijer, a head researcher and co-author of the paper, PatCID’s future development plans include expanding its database to cover chemical structures extracted from text, polymers, and Markush structures, as well as a broader selection of patents across wider date ranges. One such update to the database was newly indexed patents from all jurisdictions, which went live early in October. Dr. Meijer indicates that as the segmentation model (which extracts molecular structure images from documents) becomes more efficient and less computationally intensive, they could potentially process all patent documents from all jurisdictions, rather than limiting them to those collected with keyword selection. They are also actively working to improve their search interface to enhance user experience. As the tool’s coverage broadens, we anticipate PatCID will become a primary resource for chemical structure searches in patents.
We are excited to continue testing PatCID further and comparing its results to those of our current suite of tools, as we’re always evaluating best practices in chemical structure searching.
Have questions regarding chemical patent searching and want to discuss with our team, please contact us.
References
- Journal Article: Morin, L., Weber, V., Meijer, G. I., Yu, F., & Staar, P. W. J. (2024, August 2). PatCID: An open-access dataset of chemical structures in patent documents. Nature Communications. https://www.nature.com/articles/s41467-024-50779-y
- GitHub Repository: Morin, L. (2024). DS4SD/PatCID. GitHub. https://github.com/DS4SD/PatCID